New Collaborative Research Study Between nference and the NIH Identifies Which Mutations Enable Highly Transmissible Viruses, Like SARS-CoV-2, to Persist in Human Circulation
Published in PNAS Nexus, this study has the ability to impact future research and inform more proactive and sustainable pandemic preparedness
Excerpt from the Press Release:
CAMBRIDGE, Mass.–(BUSINESS WIRE)–nference, an AI-driven health technology company, today announced a new research partnership following the publication of a study demonstrating the value of characterizing SARS-CoV-2 viral sequences using its newly introduced ‘Distinctiveness’ metric. In collaboration with the National Institutes of Health (NIH), the study, titled “Quantifying the immunological distinctiveness of emerging SARS-CoV-2 variants in the context of prior regional herd exposure,” was published in PNAS Nexus.
“With over 13 million SARS-CoV-2 genomes sequenced globally over the last two years, there is unprecedented data to decipher how competitive viral evolution produces fitter variants,” said Venky Soundararajan, co-founder and Chief Scientific Officer of nference. “There is limited knowledge of a genomic signature that is shared primarily by the sequential dominant variants, including Omicron sublineages like BA.1, BA.2.12.1 and BA.5. The insight uncovered by the study is quite timely to begin understanding how a virus like SARS-CoV-2 is managing to adapt its genome over time to persist in human circulation for years.”
Study authors examined and compared the amino-acid changes that have occurred in all SARS-CoV-2 genomes collected to date. Results showed that new sequences accumulate mutations, which make them distinct from other viral sequences that circulated previously within the same geographical region. The study provides the first compelling genomic signal for how highly transmissive and immuno-evasive variants, like Omicron and Delta, may be able to out-compete thousands of other circulating SARS-CoV-2 lineages to emerge as dominant variants of concern within a specific region. Sequences belonging to these variants of concern exhibited significantly higher values based on the newly introduced Distinctiveness metric when compared with other contemporary sequences of that same region.
Click the button below to read the entire Press Release:
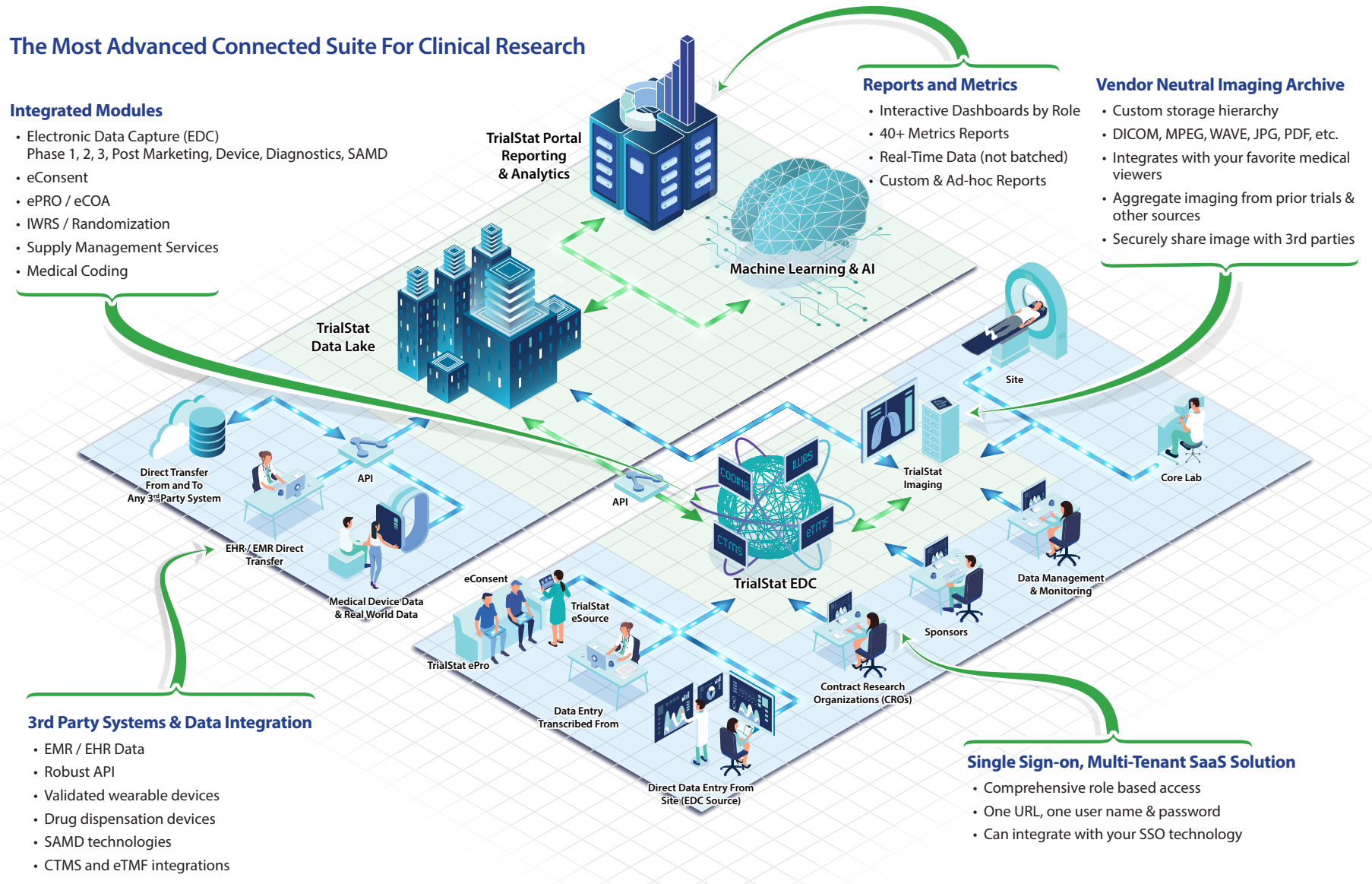
Discover What Sets TrialStat Apart From Ordinary EDC Platforms
Click the image or button below to explore our eClinical Suite Platform and discover what sets TrialStat apart from competing EDC platforms.
Request Your Demo Today!
From rapid database build through database lock, we deliver consistent quality on-time and on-budget. Ready to upgrade your eClinical toolkit?